
Our research hypothesis is: small structural variations in virulence factors modulate the virulence of different bacterial strains of the same species, and, in turn, the course of infection and disease outcome. Despite the high medical need, this variation within pathogenic species is still underexplored. Diagnostic workflows rarely distinguish between such strains and variants. Antimicrobial therapy thus currently often makes no difference between highly virulent (killer) and low-virulent (commensal or colonizer) strains. Rapid distinction of these strains would be a game-changer wherever antibiotics are used empirically (e.g. intensive care, treating the elderly), allowing for much more targeted treatment. Untargeted antimicrobial treatment also leads to the ‘blooming’ of antibiotic-resistant but clinically silent bacteria in the gut. Antibiotic stewardship based on the pathogenic potential would thus reduce unnecessary antibiotic use and the risk of antimicrobial resistance (AMR).
Our work is interdisciplinary and collaborative - microbiologists, clinicians, bioinformaticians, structural biologists, cell biologists and bioengineers work on different aspects of bacterial strains and variants. Or current focus are bacterial adhesins, the virulence factors that are essential for first contact with the infected host, and for establishing and maintaining an infection by attaching to different cells and surfaces.
We are recruiting 15 fully-funded doctoral candidates for 36-month positions across our European network, with guaranteed secondments in both academic and industrial partner institutions.
The StraDiVarious research programme comprises four integrated work packages that progress from genomic strain characterisation to diagnostic device development. WP1 establishes the strain variant collection and adhesome profiles. WP2 investigates adhesin-mediated host interactions using organ-on-chip models. WP3 characterises adhesin-receptor structures at atomic resolution. WP4 develops rapid biosensor devices for clinical applications. Fifteen doctoral candidate projects are distributed across these work packages, ensuring comprehensive training in complementary methodologies.
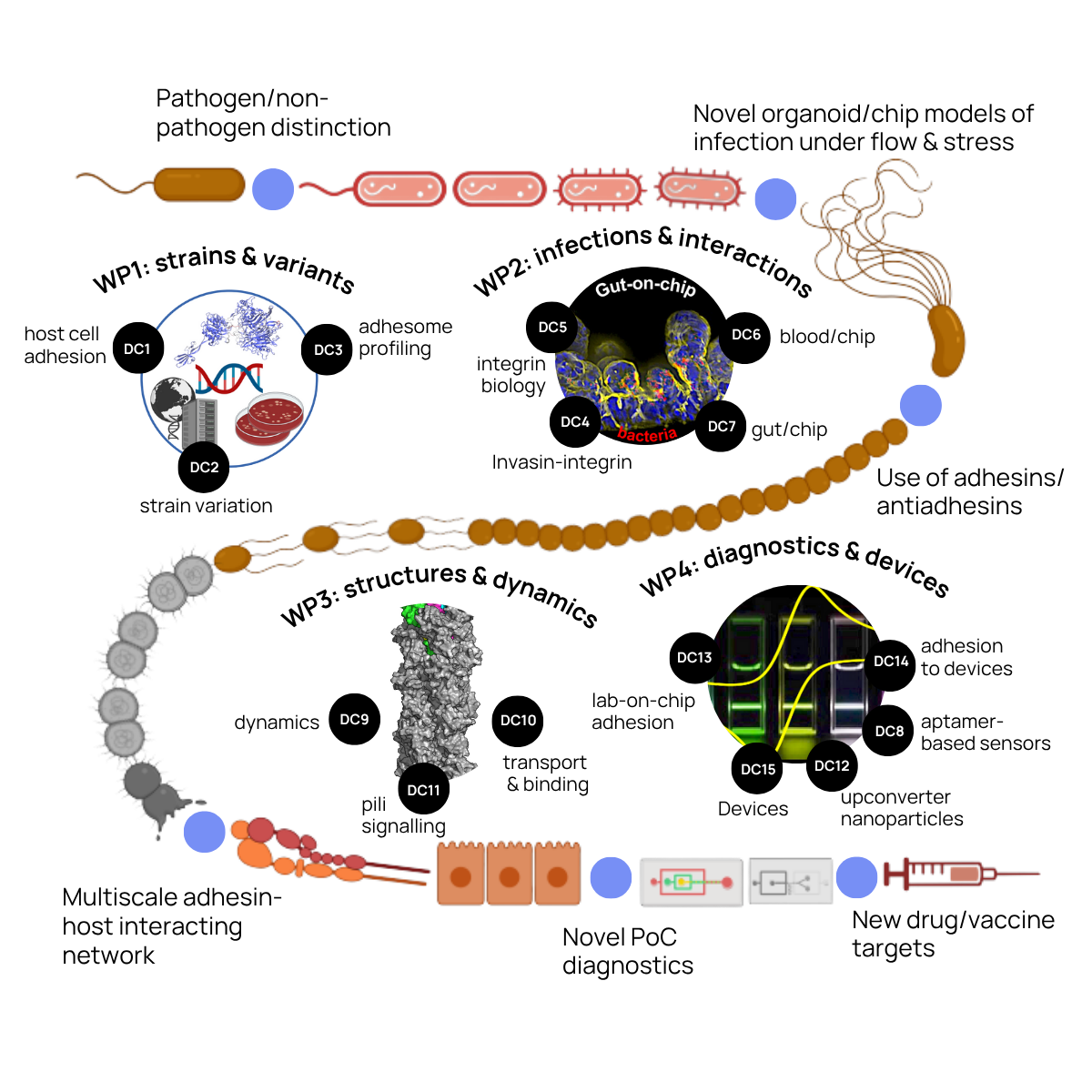
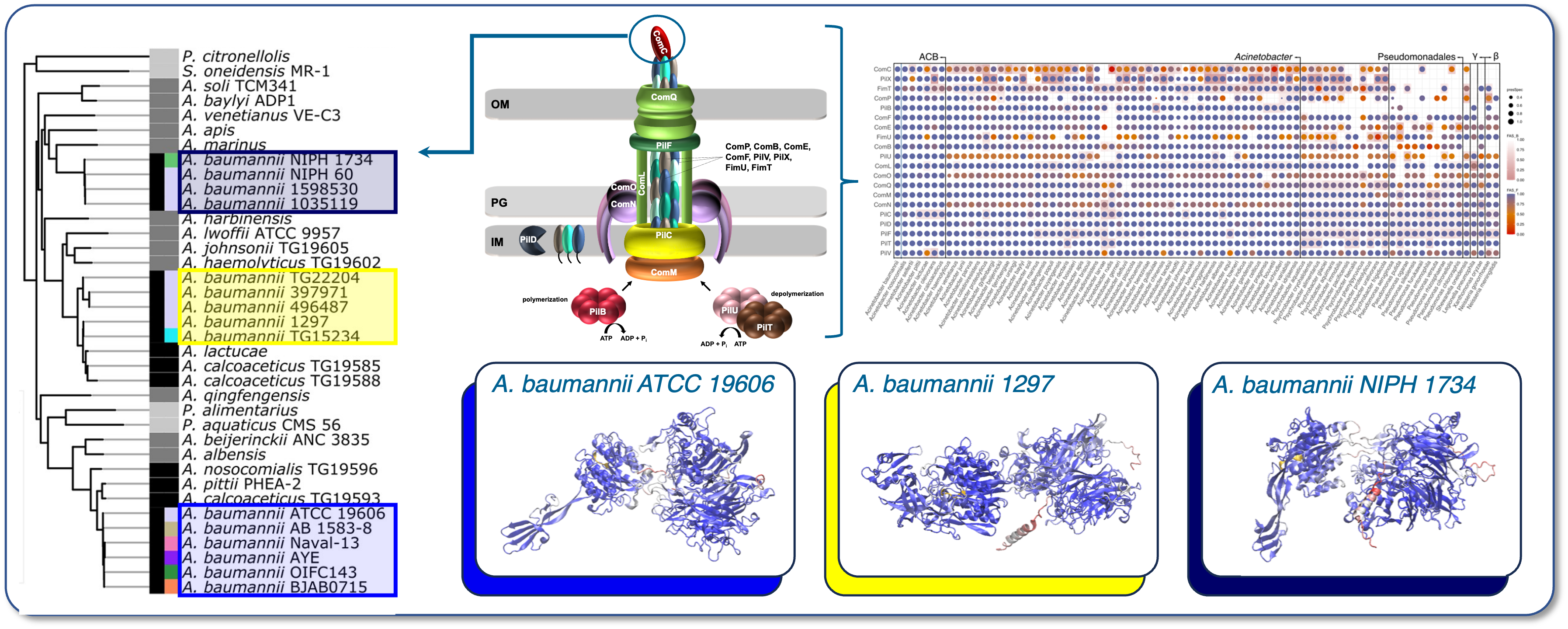
Establishing the StraDiVarious Variant Collection (StraVaC), characterising species-specific adhesomes, and using machine learning to predict pathogen virulence based on adhesin variation.
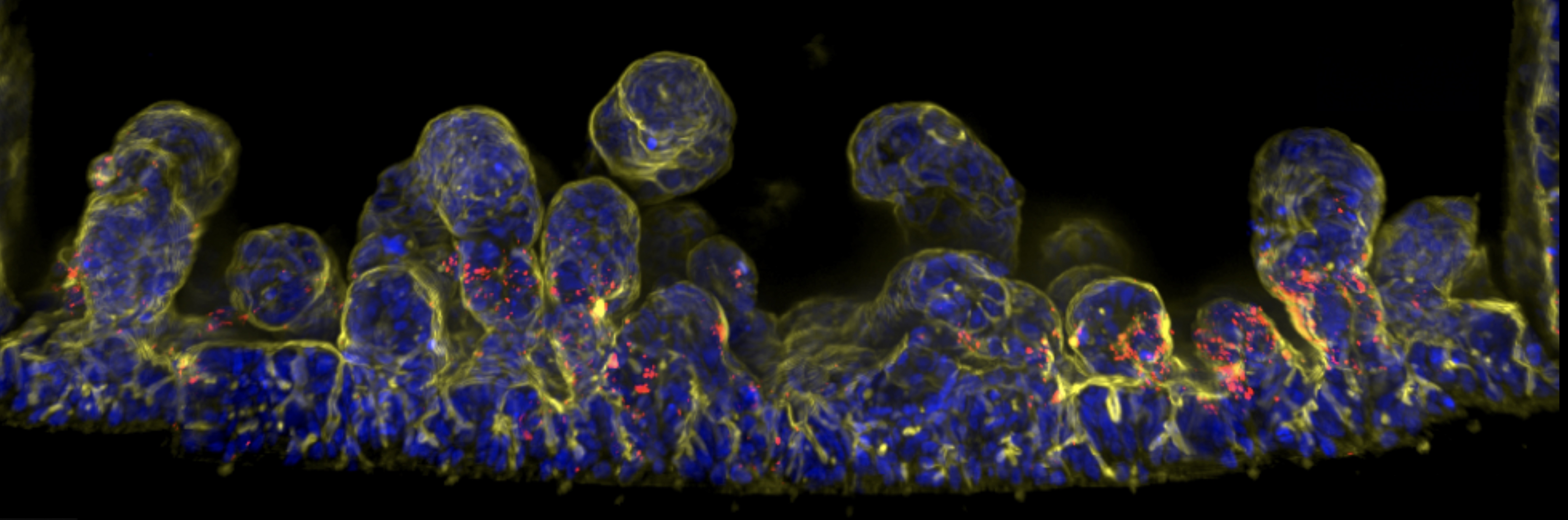
Developing organ-on-chip models to study adhesin-mediated bacterial adhesion, colonisation, and host cell manipulation, reducing dependence on animal models.
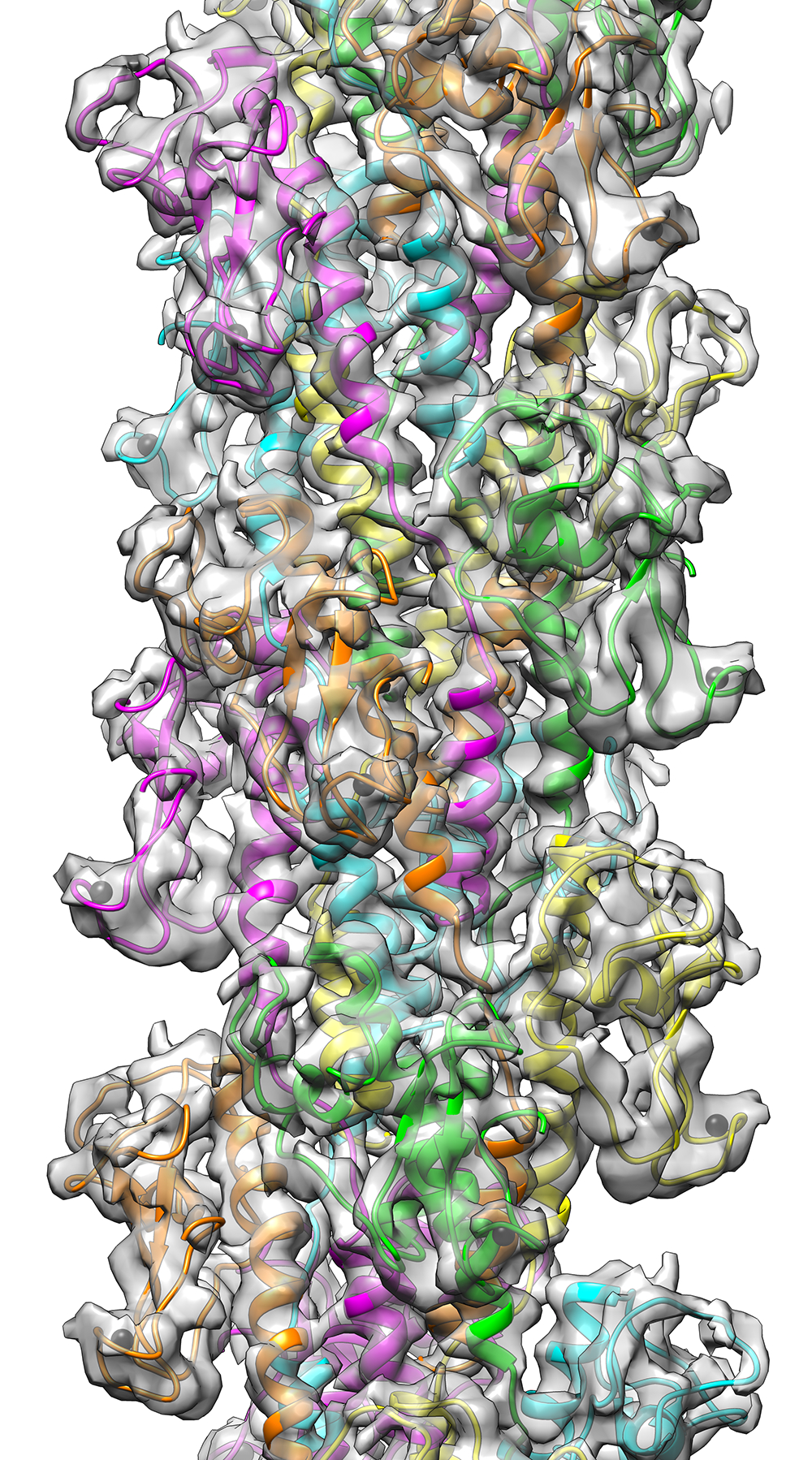
Performing atomic-scale structural characterization of adhesin-host receptor interactions using cryo-EM, EPR, NMR, and integrative modelling to identify drug targets.
Developing rapid biosensors and diagnostic devices using nanoparticles, electrochemical biosensing, and aptamers to detect and measure virulence traits for commercialisation.
Reducing the global burden of AMR requires action at the research, local, government and international level. Join our team of dedicated researchers.