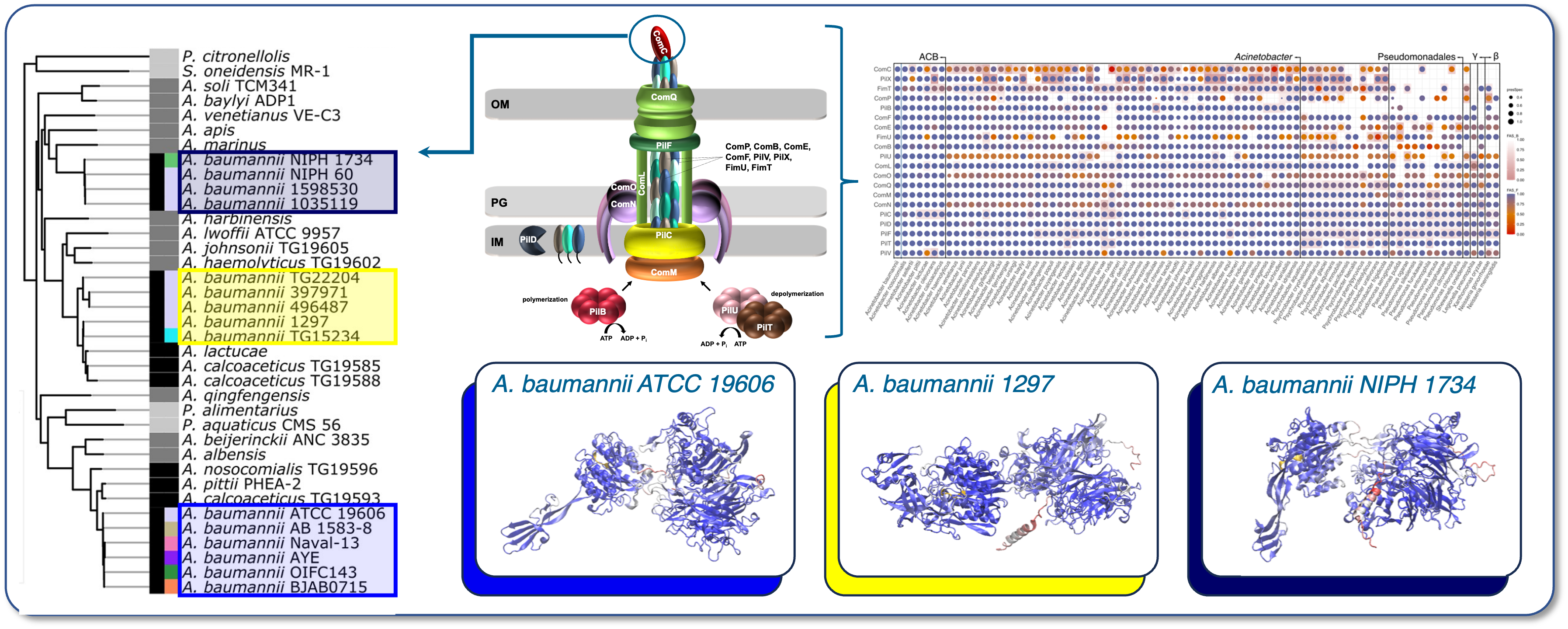
Closely related bacterial strains can differ dramatically in their ability to cause disease. A major factor behind this variation lies in adhesins—surface proteins that mediate binding to host tissues and initiate infection. This work package investigates how adhesin diversity influences bacterial virulence and aims to identify molecular features that distinguish high- and low-virulence strains.
We will establish the StraDiVarious Variant Collection (StraVaC), a curated resource linking ESKAPE pathogen genomes to phenotypic and clinical data on infection severity. Using comparative genomics, we will analyse the evolution and diversity of adhesomes, the complete set of adhesins expressed by a bacterial strain, within and across species. A key method will be protein feature architecture-aware phylogenetic profiling, allowing us to detect structural and evolutionary changes in adhesins associated with virulence. By integrating adhesome profiles with virulence data, we will use machine learning to identify features predictive of pathogenicity. Selected strains - natural variants and mutants of A. baumannii, P. aeruginosa, and S. aureus - will be analysed for biofilm formation and host cell adhesion. Quantitative proteomics will reveal strain-specific differences in adhesin expression and identify host receptors involved in adhesion, invasion, and immune evasion. To study the functional relevance of key variants, we will collaborate with experimental partners from the consortium using organ-on-chip systems and 3D human tissue models that replicate host environments and responses. Together, these approaches will provide an integrated understanding of how adhesin variation shapes bacterial virulence and support the development of genome-based tools for infection prediction and control.

We are recruiting 15 fully-funded doctoral candidates for 36-month positions across our European network, with guaranteed secondments in both academic and industrial partner institutions.
We are recruiting 15 fully-funded doctoral candidates for 36-month positions across our European network, with guaranteed secondments in both academic and industrial partner institutions.