The StraDiVarious research programme is organised into four integrated work packages that span the complete research pipeline from strain characterisation to diagnostic development. WP1 establishes the bacterial strain variant collection and characterises adhesomes. WP2 investigates adhesin-mediated host cell interactions using advanced organ-on-chip models. WP3 performs atomic-scale structural characterisation of adhesin-receptor complexes. WP4 develops rapid biosensor devices and diagnostic technologies for commercialisation.
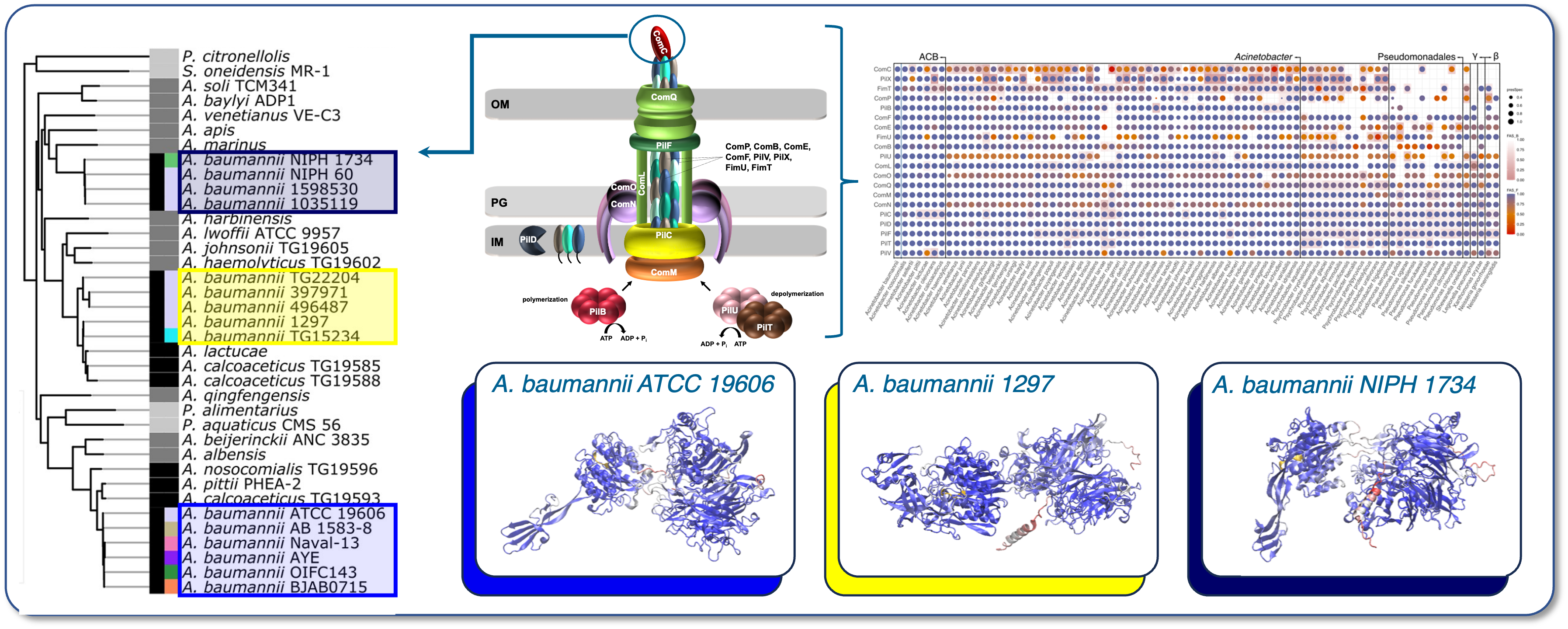
Closely related bacterial strains can differ dramatically in their ability to cause disease. A major factor behind this variation lies in adhesins—surface proteins that mediate binding to host tissues and initiate infection. This work package investigates how adhesin diversity influences bacterial virulence and aims to identify molecular features that distinguish high- and low-virulence strains.
We will establish the StraDiVarious Variant Collection (StraVaC), a curated resource linking ESKAPE pathogen genomes to phenotypic and clinical data on infection severity. Using comparative genomics, we will analyse the evolution and diversity of adhesomes, the complete set of adhesins expressed by a bacterial strain, within and across species. A key method will be protein feature architecture-aware phylogenetic profiling, allowing us to detect structural and evolutionary changes in adhesins associated with virulence. By integrating adhesome profiles with virulence data, we will use machine learning to identify features predictive of pathogenicity. Selected strains - natural variants and mutants of A. baumannii, P. aeruginosa, and S. aureus - will be analysed for biofilm formation and host cell adhesion. Quantitative proteomics will reveal strain-specific differences in adhesin expression and identify host receptors involved in adhesion, invasion, and immune evasion. To study the functional relevance of key variants, we will collaborate with experimental partners from the consortium using organ-on-chip systems and 3D human tissue models that replicate host environments and responses. Together, these approaches will provide an integrated understanding of how adhesin variation shapes bacterial virulence and support the development of genome-based tools for infection prediction and control.
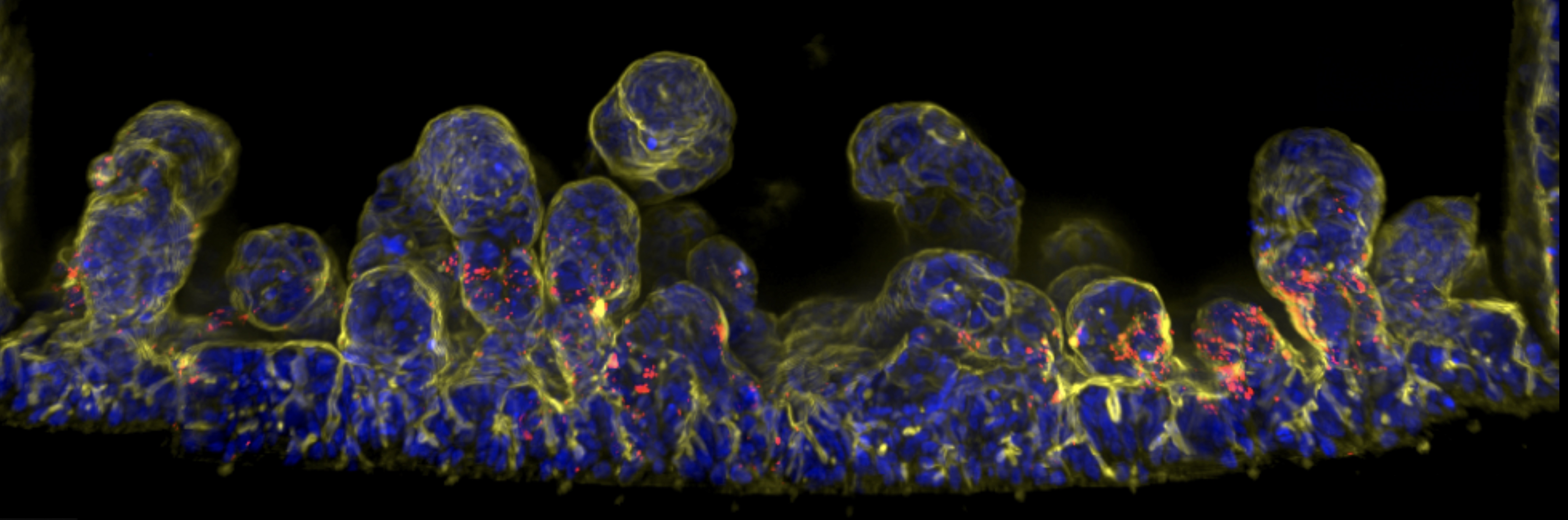
We aim to understand binding of pathogen variants to host cells, how variants manipulate host cell responses, and to develop innovative organ-on-chip models to study strain and variant adhesion/colonisation of the host. We will use a combination of in vitro assays, cell lines, human cells and tissues, and organ-on-a-chip models, to understand how pathogenic variants interact with host cells (adhesion, colonization) and manipulate their responses (cell signaling, inflammation).
There is an urgent need to develop laboratory models of humanised organs to reduce dependence on animal models in scientific research. Animal models are expensive, time-consuming, ethically problematic, and often poor predictors of human biological responses. To address this, we aim to create next-generation humanised models of the gut and vasculature using advanced organ-on-chip technology to study bacteria-host interactions. This technology enables more accurate modelling of human tissues and organ-level structures compared to traditional cell culture methods. By using organ-on-chip (OoC) systems, we can observe and control these interactions with unprecedented precision, offering new insights into the mechanisms of colonisation and infection that were previously unattainable. Gut-on-chip models will be developed at Institut Pasteur, using patients derived intestinal organoids maturated in OoC (Institut Pasteur, France) to analyse the dynamics of bacterial adhesion, colonisation and invasion. Human vasculature-on-chip models will be developed at Heriot-Watt University, Edinburgh and used to investigate mechanisms of bacterial blood infection and colonisation alongside collaborators at Lund University.
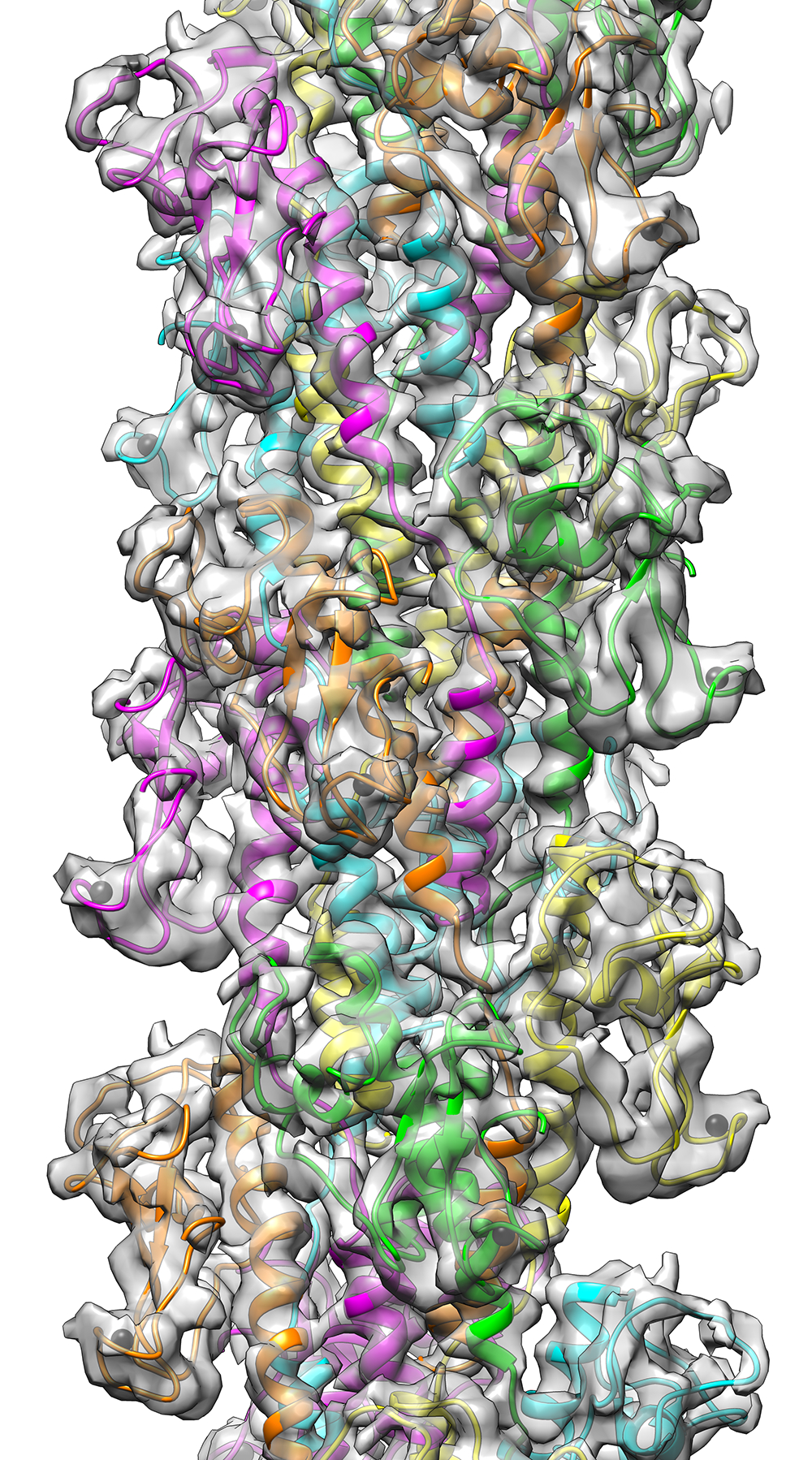
We will perform structural characterization of adhesin-host receptor interactions at an atomic scale, and use this information to understand differences between high-virulent and low-virulent bacteria. This will also allow for identification of potential drug targets.
Development of new diagnostic, anti-adhesion and antimicrobial strategies requires a comprehensive understanding of adhesin-host receptor binding and associated signalling events. To achieve this, we focus on the interaction networks of key bacteria-host protein complexes, and on the effect of small variations in virulence factor sequence on structure and function. We will study the dependence of adhesin sequence length and repeat number on the conformational dynamics (bending, twisting) of the molecule. This will be achieved by combining high-resolution structural biology methods such as cryo-EM and structural EPR with different modelling approaches. Similarly, we will study the interactions and interaction networks of key adhesins from ESKAPE pathogens using a combination of NMR, cryoEM and integrative modelling. CyroET of adhesins in different conditions and with different host cell models will be used to test adhesion capacity of pili and develop anti-adhesion strategies.
We will develop rapid, innovative biosensors for diagnostics and advanced bioassay systems to study how host cells bind and respond. We will then use these tests to explore links with antimicrobial susceptibility testing (AST).
Our bioassays and diagnostic devices will be based on the targets and interactions identified and characterised for ESKAPE pathogens. The aim is to quantify interactions and develop rapid tests and test kits to detect, screen, and measure virulence traits. Sensor binding assays will be developed based on novel methodologies such as recently developed nanoparticle labels and, emerging materials and approaches for electrochemical biosensing and stable recognition elements such as aptamers and affimers. Diagnostics flow cartridges and spheroid-on-a-chip devices will be designed and tested and the optical and electrochemical sensing approaches will be integrated to obtain dynamic systems to quantify binding. The ultimate aim is to generate diagnostic devices for commercialisation.
Fifteen fully-funded doctoral candidate positions are distributed across our European network, each focusing on specific aspects of bacterial adhesion research. Projects span molecular microbiology, structural biology, bioinformatics, organ-on-chip technology, and diagnostic device development. All candidates receive interdisciplinary training through secondments at both academic and industrial partner institutions over 36 months.
To find out about more, click the projects below and choose your preferred one.
The PhD candidate will study bacterial adhesins and their interactions with host receptors. The work centres on adhesins from Klebsiella and Enterobacter species, that are members of the ESKAPE pathogens notorious for their emerging multidrug resistance.
The research includes the genomic mining for potential pathogenicity factors differentiating "real world" patient infecting "colonizing strains" (with low virulence) from "killer strains" (with high virulence). These strains will be made available from the routine diagnostic laboratories of the institute. Genomes will be sequenced (using Nanopore-technology) and genome analysis will be performed in collaboration with Prof. Ebersberger. The respective genes will be genetically characterized, recombinantly expressed (e.g., in E. coli) or deleted in the respective patient isolates. Virulence will be determined in infection assays analysing bacterial binding to matrix proteins or in host-cell or invertebrate infection models. Potential binding partners will be analyzed in collaboration with the consortium with a special emphasis on integrins.
Methods will include the generation of bacterial mutants and other methods in molecular microbiology, protein expression and purification, and both cellular infection assays and adhesion assays to matrix proteins. An integral part of the work is the identification of host cell receptors using, e.g., CRISPR-Cas9 technology. For microscopic analysis, a brand-new Zeiss Apotome microscope is available allowing real-time microscopy of infection processes.
The work will be done in close collaboration with groups in the network interested in cellular models, bacterial adhesion, bioinformatics and in anti-virulence drug development. The position thus offers the opportunity of internships with both academic and industrial partners.
The PhD candidate will study bacterial cell-wall and secreted virulence factors involved in host adhesion and immune evasion. The work centers on virulence factors from Staphylococcus aureus and Pseudomonas aeruginosa, which are ESKAPE pathogens notorious for their biofilm-forming capabilities and their emerging multidrug resistance. In this project, you will identify and characterize the host interactions of these species using methods in quantitative and structural proteomics. The research questions span from characterizing the interactomes of intact bacteria to that of isolated virulence factors and their domains. Using infection assays, you will establish the importance of selected adhesins in different stages of the infection process and identify host cell receptors. The work will be done in close collaboration with groups in the network interested in cellular models for host adhesion, biofilm formation and bioinformatics. The position offers the opportunity of internships with both academic and industrial partners, including the characterization of pathogen adhesion to host cells using a vasculature-on-chip model, and the adhesion of biofilm forming bacteria to biomaterials.
Methods will include quantitative affinity-purification and surface adsorption mass spectrometry using isolated virulence factors and intact bacteria, respectively. Structural proteomics (cross-linking and hydrogen-deuterium exchange mass spectrometry) will be used to map the host-pathogen interface at high-resolution, optionally followed by structural determination of the complexes using cryo-EM. These methods are complemented with bioinformatics and structural modeling and docking. Other methods applied include standard methods in molecular microbiology, as well as cellular infection and biophysical adhesion assays.
Understanding why some bacterial strains are more virulent than others—both across and within species—is a central challenge in infectious disease biology. Genomic differences underpin much of this variability, but pinpointing which variations matter, and why, remains unresolved.
In this PhD project, you will investigate how genomic variation influences virulence, focusing on bacterial adhesins—key proteins mediating host-pathogen interactions. Using a multi-scale bioinformatics approach, your goal will be to map the composition and diversity of the adhesome (the full complement of adhesins) across multiple bacterial pathogens, and to correlate this diversity with virulence phenotypes. Ultimately, your project will uncover how adhesome variability contributes to pathogen virulence.
You will integrate genomic and phenotypic data (in collaboration with DC1) to build the StradiVarious Variant Collection, a comprehensive database of bacterial strains with varying virulence profiles. On this basis, you will investigate the evolutionary trajectories of virulence-associated proteins, especially adhesins, across diverse bacterial genomes. You will explore and chart inter- and intraspecific variation in the adhesome using feature-architecture-aware phylogenetic profiling and pan-genome graphs. On this basis, you will identify adhesin orthologs with altered structures, evolutionary histories, or domain architectures as potential drivers of altered bacterial virulence. You will apply supervised and unsupervised machine learning to correlate adhesin variation with virulence and generate a shortlist of candidates for experimental validation in partner labs.
The PhD candidate will study bacterial adhesins and their interactions with host receptors. The work centers on adhesins from Klebsiella and Enterobacter species, which are two of the ESKAPE pathogens notorious for their emerging multidrug resistance. In this project, you will help to identify bacterial adhesins in the genomes of ESKAPE strains and generate knockout mutants. Using infection assays, you will establish the importance of these adhesins in different stages of the infection process and identify host cell receptors.
The work is in close collaboration with other groups in the network interested in host cell receptors, and in anti-adhesion drug development. The position thus offers the opportunity of internships with both academic and industrial partners.
Methods will include the generation of mutants and other methods in molecular microbiology, protein expression and purification, and both cellular infection assays and biophysical adhesion assays. An integral part of the work is the identification of host cell receptors using mass spectrometry.
In this project, the PhD candidate will study cell biological responses to adhesin-integrin interactions, and the influence of adhesin variants on cell responses. The candidate will investigate adhesin variants from K. pneumoniae, Enterobacter and other ESKAPE pathogen strains, focussing particularly on invasins. Interactions of adhesins and their variants with host cell integrins will be studied by cell adhesion assays and affinity assays. The candidate will elucidate how pathogen adhesin binding to integrins on the host cell regulate inflammatory responses, eg cell signalling, cytoskeletal organization, gene expression and functional responses in the cells. Main techniques to be used will be biochemical assays, microscopy, flow cytometry, qPCR and RNAseq approaches. This project will generate an understanding of how interactions between bacteria and their host cells regulate signalling and inflammation responses. The project will be conducted in close collaboration with both Academic and Industrial partners in the network through internships.
In this project, the PhD candidate will develop an organ-on-chip device to support the growth and stability of 3D human microvasculature to study pathogens responsible for blood infections, in particular ESKAPE pathogen Staphylococcus Aureus in collaboration with researchers at Lund University, Sweden. The humanised endothelial-on-chip model will be used to study bacterial adhesion, and host colonisation in blood vessels that mimic in vivo conditions, both in the biology and the biophysical flow characteristics. The candidate will study conditions in which pathogens adhere to host cells, and how colonisation progresses in real-time. The response of the host cells to infection will also be studied, and may shed light on new targets for therapeutic intervention. This project will align strongly with the 3Rs by developing advanced in vitro platforms towards the replacement of animal models in research.
Methods will include microfluidic design, fabrication and characterisation using a combination of microscopy, image analysis and flow analysis. Cell culture and advanced fluorescence microscopy to study 3D vasculature network formation and biological assays for assessing host cell interactions with pathogens.
The PhD student will work on the impact of the microenvironment of the human gut on colonisation and virulence of enteric bacterial pathogens (EHEC). In this project you will use a highly physiological human colon model, combining patient-derived colonic organoids with organ-on-chip (OoC) and enabling to modulate the key gut environmental cues: mechanical stimulation (peristalsis and shear stress) and microbiota metabolites. Using several EHEC strains based on their differences in adhesins or Type IV pili, you will help to characterize the colonization, adhesion of these variants, and the consequence of the gut environmental cues on the EHEC virulence.
The work is in close collaboration with other groups in the network interested in host cell receptors, and in anti-adhesion drug development. The position thus offers the opportunity of internships with both academic and industrial partners.
Methods will include the generation of human colonic organoids, barrier maturation in OoC, infection assays in OoC modulating mechanical forces and microbiota metabolites, live and fixed imaging quantifying the bacterial adhesion and colonization, CFU counting, barrier leakage assays, immunofluorescence and RTqPCR analysing the downstream impact of infections on the colonic barriers.
The PhD candidate will study aptamers specific to Pseudomonas aeruginosa towards the design of novel diagnosis systems. The work focuses on detecting Pseudomonas aeruginosa, that is an ESKAPE pathogen and well-known by its antibiotic resistance. In this project, the PhD candidate will select relevant ligands specific to this pathogen which are key to develop improved detection systems against its variants and that could further inform therapeutic strategies.
The work is a close collaboration with other groups in the network interested in the development of devices and diagnostics of virulence via adhesin-host binding. The position thus offers the opportunity of internships with both academic and industrial partners.
Methods will include constructing heterologous Escherichia coli strains expressing P. aeruginosa adhesins. These will be then used to select high-affinity aptamers against P. aeruginosa adhesins through cell-SELEX. Aptamers affinity and selectivity will be characterized and further used to develop electrochemical aptasensors. Clinical isolates will be used to establish the aptasensor readout.
For the Stradivarious project the PhD candidate will be based at The University of Manchester, United Kingdom, and will be exploring the conformational flexibility and structural architecture and ensemble of adhesins in host-ESKAPE interactions. 'ESKAPE' pathogens top the World Health Organisation's global bacterial pathogen priority list. Target adhesins will be isolated and structurally characterised. Their interactome and dynamics will be then studied in presence/absence of host cells, by state-of-the-art methods, such as PELDOR spectroscopy and cryo-EM. This will enable the dissection of host-ESKAPE interactions at the molecular level and identify the key proteins in these fundamental cellular processes.
The main objectives of the project will be to investigate adhesin biomechanics, drug-protein and protein-protein interactions under infectious conditions and in pathogen-derived variants.
The PhD candidate will be trained on and use state-of-the-art structural biology approaches, such as Electron Paramagnetic Resonance (EPR) spectroscopy and cryo-Electron Microscopy (cryo-EM), combined with modelling and Molecular Dynamics (MD) simulations.
There are no accurate models for how trimeric autotransporters (TAAs) bind host effectors; all of the studies so far have involved mutagenesis and foot-printing. We aim to rectify this gap. AlphaFold does a poor job of modelling these structures and complexes (1) because they have unusual sequences and (2) because they aren't globular.
The project involves studying TAAs from ESKAPE pathogens (eg. Acinetobacter) to determine how they bind host effectors (collagen, laminin etc) via cryoEM and structural EPR spectroscopy (with Christos Pliotas, University of Manchester). The student will then connect this information to strain and variant differences/changes in virulence, as this will lead to improvements in diagnostics. We will then extend this work to model how invasins, related proteins, from at least one ESKAPE pathogen studied in StraDiVarious (K. pneumoniae and E. cloacae) bind their targets. Finally, we will model how the inside-out barrels insert into the membrane by studying the (BAM-invasin complex), building on work by our collaborator (Dirk Linke, University of Oslo). The project will allow both academic and industry secondments, using state of the art structural biology equipment.
Methods will include protein expression and purification of membrane proteins, as well as EPR spectroscopy (in Manchester) and cryoEM, both locally and on secondment. Modelling the complex with the latest tools and the validation of those models through mutagenesis will also be a key part of the techniques used.
This PhD project will investigate the structural basis of bacterial pili-mediated adhesion, a critical process in host colonization and biofilm formation. Using an integrative structural biology approach combining cryo-electron microscopy, NMR spectroscopy, and advanced molecular modeling, the research will unravel the molecular architecture and conformational dynamics of pili and their interactions with host receptors. Complementary biophysical and functional assays will be employed to correlate structure with adhesion mechanisms, providing insights into how bacterial pili assemble, extend, and mediate binding under physiological conditions. The ultimate goal is to establish a detailed structural framework for understanding pili-driven adhesion, opening avenues for the development of anti-adhesion strategies to combat infections.
This project includes two mandatory secondments: 1-Three months in the first year at the Lund University (Sweden) to identify host or bacterial protein partners in interaction with pili by proteomic Mass Spectrometry analysis of extracted pili/whole bacteria. 2-Three months in Y3 with the industrial partner Corti (Norway) to develop and test anti adhesion strategies based on structural data.
This project will explore how novel upconverting nanoparticles (UCNPs) can be used to detect and measure the binding of bacterial virulence factors. Unlike traditional fluorescent dyes, UCNPs are highly stable, resist photobleaching, and operate in the near-infrared range - allowing deeper tissue penetration for more reliable detection. These properties make them ideal for developing next-generation diagnostic tools.
You will gain hands-on experience in nanoparticle synthesis and characterisation, surface functionalisation with biomolecules, and the design of simple optical detection systems to track binding events. As the project progresses, you will evaluate assay performance (sensitivity, specificity, limit of detection) and test rapid-readout formats, including fluidic flow-cell systems that provide real-time binding data.
The project also includes international secondments: at Lund University, you'll explore applications with host/microbe interactions. You will also have a secondment to develop skills in spheroid and organelle culture and while at Selex in the Netherlands, you'll gain experience on translating technologies into real-world diagnostic settings. These secondments might change depending upon how your project develops and the directions that you take.
In the Pamme Research Group we are designing microfluidic lab-on-a-chip devices for clinical diagnostics as well as biomedical research, environmental analysis and the synthesis of smart materials.
In this project we will develop electroanaytical sensor devices and study how host cells bind and respond. We will design microfluidic flow cells and integrate them with printed electronics to created bioelectroanalytical sensors. With these sensors we will study adhesion-cell and protein-protein interactions. We will immobilise target integrins and receptors on the electrode surfaces and monitor adhesion interactions under the highly controlled conditions achievable within microfluidic environments, i.e. controlling concentrations in space and time and controlling shear stress in space and time. We will work across the doctoral training network to implement molecular binding elements such as whole cells, proteins and newly developed aptamers. This will allow us to investigate the variance in adhesion between different pathogens.
We will develop methods and apply in electroanalytical measurement techniques to study binding events between immobilised recognition elements on the electrode surface and the surrounding medium. This will involve various electrode materials and immobilisation strategies. The electrodes will be integrated into microfluidic flow cells, which we will design and fabricate, first for single readout, then for parallelisation and simultaneous readout.
The PhD candidate will develop bacterial biofilm models with single or multiple bacterial species under aerobic and anaerobic conditions. You will use these model biofilms to test medical implants and other medically relevant surfaces. They will also be used to test the effect of mutations in surface attachment genes of individual, implant-related ESKAPE pathogens.
The work is a close collaboration with an Oslo-based company interested in medical implants, Corticalis AS. The position thus offers the opportunity to work in an academic setting but in close collaboration with a commercial partner.
Methods will include microbial growth of pathogenic bacteria on surfaces under different growth conditions, including the use of fermentation equipment and anaerobic chambers. Analytical methods will include (but are not restricted to) light microscopy/FISH, molecular biology (qPCR), and possibly next-generation sequencing techniques.
The PhD candidate will assist in the development of a direct from sample diagnostic system that differentiates "killers" and "colonisers". The work will centre on the diagnosis of bacterial infections, where distinction between pathogens and colonising, contaminating bacterial flora is key. In this work consecutive and optimised versions of the instrument and the biochemical assays will be tested in real life clinical settings. From a more fundamental perspective, diagnosis of other infectious syndromes will be included. In addition, expertise on adhesin biology as available in the Stradivarious network will be used to optimise and refine our diagnostic tests. The study of bacterial enrichment through adhesin and ligand affinity assays will be piloted.
The work is in close collaboration with other both academic and industrial clinical microbiology and infectious diseases R&D groups in and outside the Stradivarious network. The position offers the opportunity of internships with both academic and industrial partners.
Methods will include the experimental clinical validation and verification of new generations of the KAIROS diagnostic platform. Clinical studies will be performed in various healthcare settings, allowing new applications of the platform to be developed.
We are recruiting 15 fully-funded doctoral candidates for 36-month positions across our European network, with guaranteed secondments in both academic and industrial partner institutions.